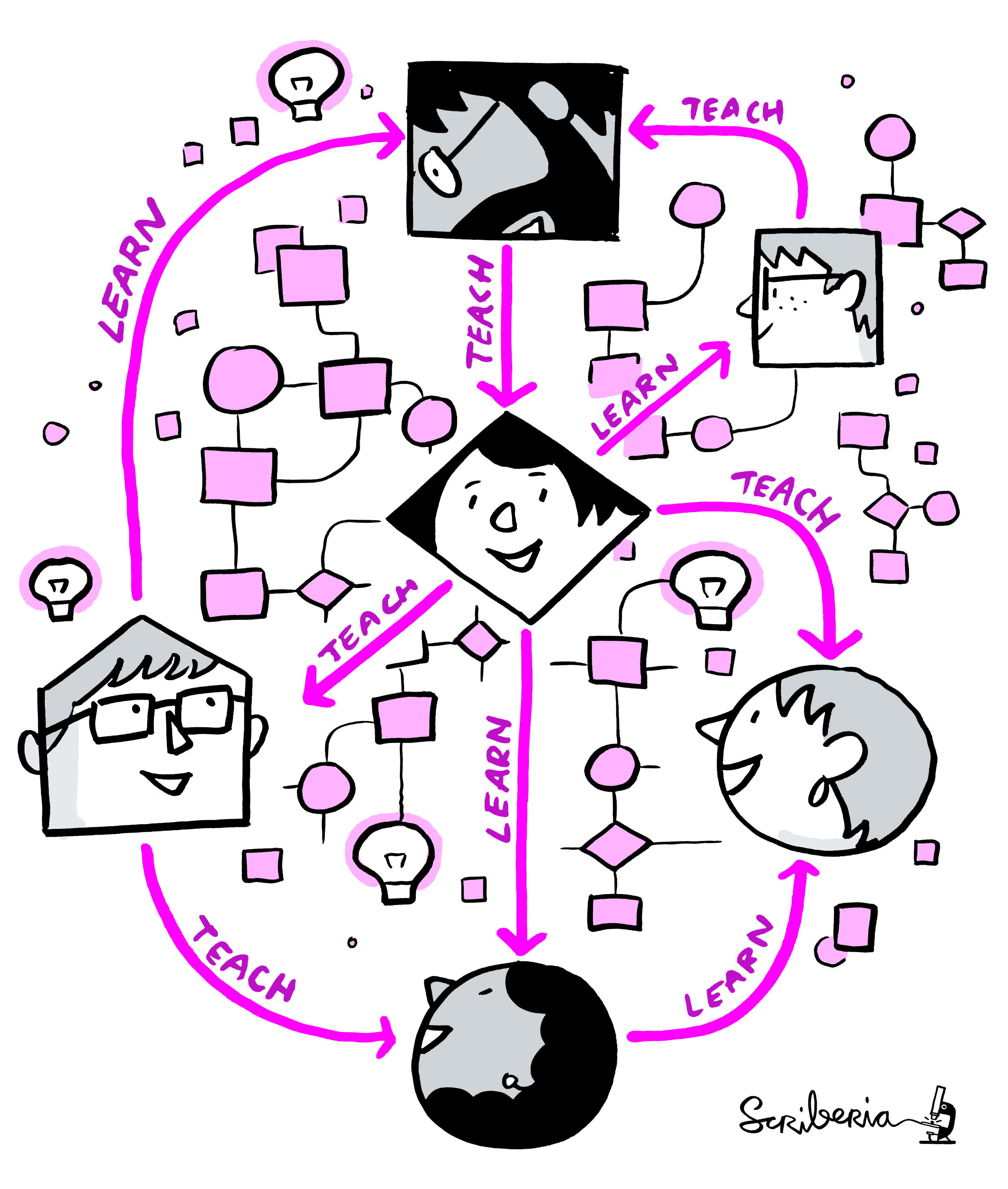
Welcome to the Mannheim Open Science Day Reprohack

Thank you ! 🙏
University Of Mannheim Open Science Office!
https://www.uni-mannheim.de/open-science/open-science-office/

Especially:
Philipp Zumstein ✨
David Morgan ✨
Agenda
| Time | Event |
|---|---|
| 10:00 | Welcome and Orientation |
| 10:10 | Ice breaker session in groups |
| 10:20 | TALK: Joel Nitta: ‘Reproducible analyses with targets and docker: An example from ReproHack’ |
| 10:45 | Anna Krystalli: ‘Tips and Tricks for Reproducing and Reviewing.’ |
| 11:10 | Select Papers, Chat and coffee |
| 11:30 | Round I of ReproHacking (break-out rooms) |
| 12:30 | Re-group and sharing of experiences |
| 12:45 | LUNCH |
| 13:45 | Round II of ReproHacking (break-out rooms) |
| 14:45 | Coffee break |
| 15:00 | Round III of ReproHacking (break-out rooms) - Complete Feedback form |
| 16:00 | TALK: Camille Landesvatter: Writing reproducible manuscripts in R Markdown and Pagedown |
| 16:25 | Re-group and sharing of experiences |
| 16:50 | Feedback and Closing |
House Keeping:
Toilets: Downstrairs one level
WiFi: uni-event
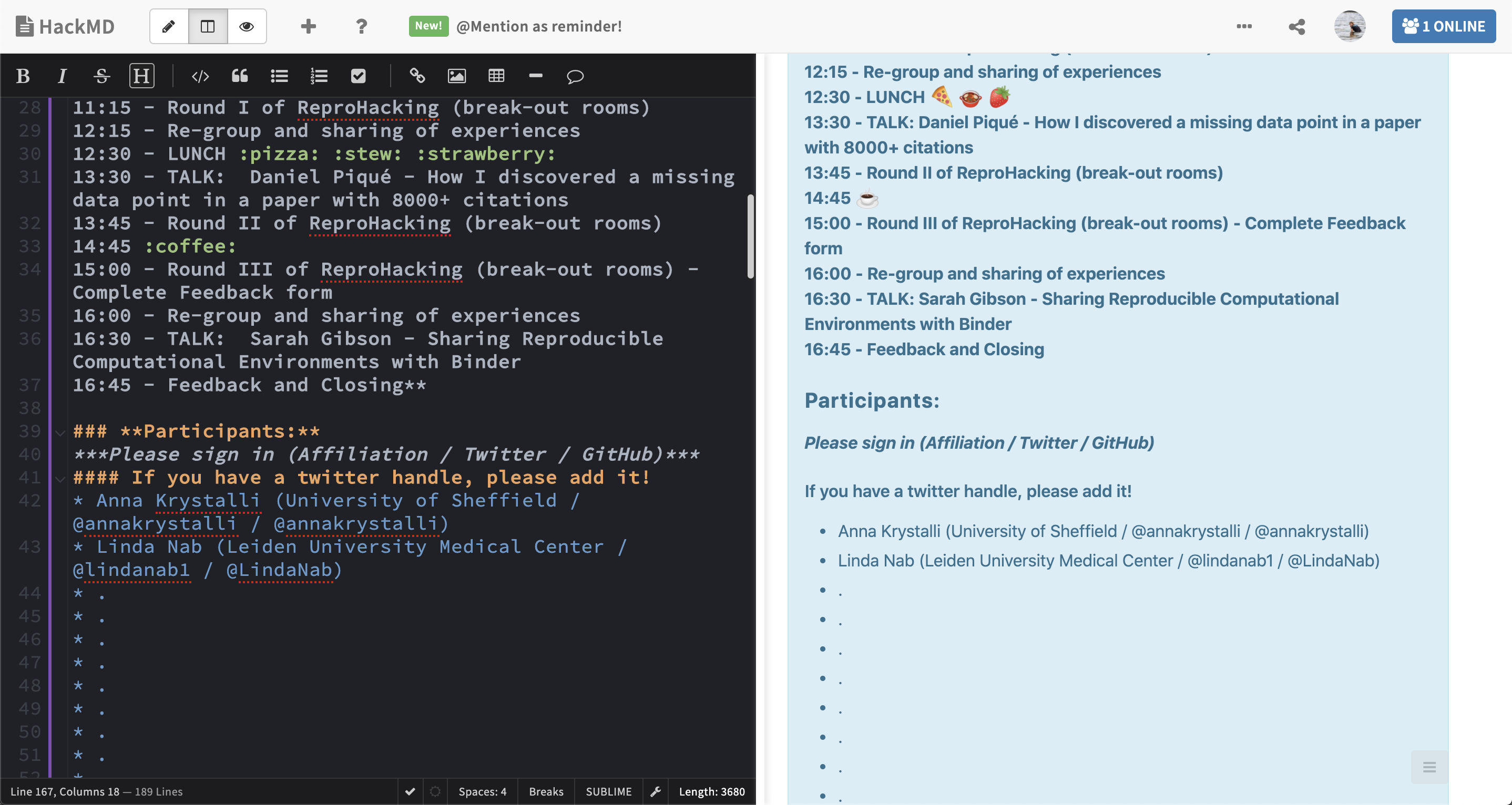
ReproHack2022Want to join in on Joel’s Live demo? You’ll need to install Docker! docs.docker.com/get-docker/
ReproHack hackpad ➡️ hackmd.io notepad
Ice breaker: Introductions
Who am I?
Dr Anna Krystalli (@annakrystalli)
Research Software Engineering Consultant
r-rse2019 Fellow Software Sustainability Institute
Software Peer Review Editor rOpenSci
Core Team Member ReproHack
Why am I here?
I believe there’s lots to learn about Reproducibility from working with other people’s materials and engaging with real published research code and data.
Who is my favorite animated character?
Stitch!
Your turn
Who are you?
Why are you here?
Who is your favorite animated character?
TALK
📢 Joel Nitta
Project Research Associate in the Iwasaki Lab at the University of Tokyo.
‘Reproducible analyses with targets and docker: An example from ReproHack’
Tips for Reproducing & Reviewing
ReproHack Objectives
Practical Experience in Reproducibility
Feedback to Authors
Think more broadly about opportunities and challenges
Additional Considerations
Reproducibility is hard!
Submitting authors are incredibly brave!
Thank you Authors! 🙌
Without them there would be no ReproHack.
Show gratitude and appreciation for their efforts. 🙏
Constructive criticism only please!
🔍 Reproducing & Reviewing

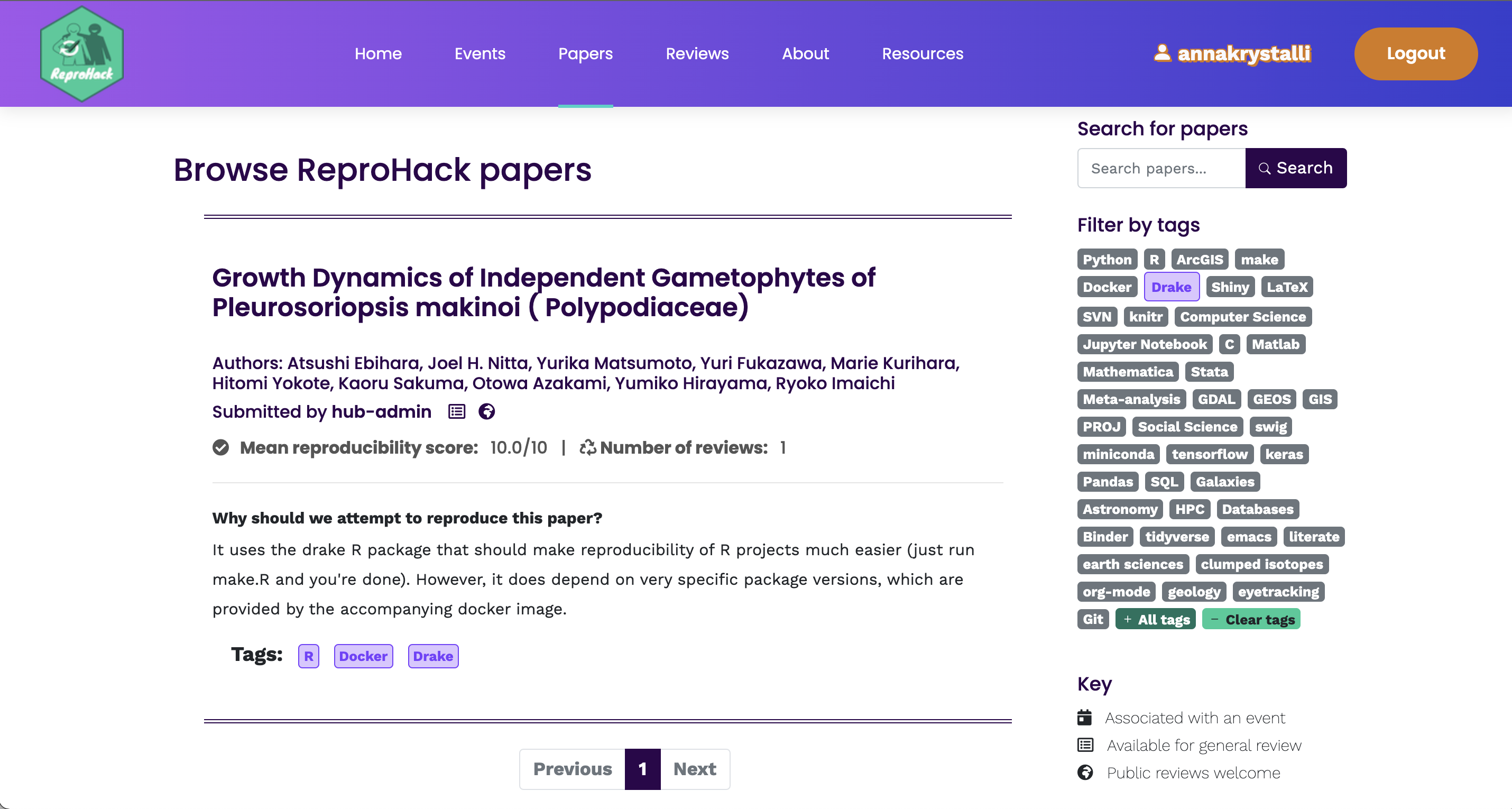
Selecting Papers
Information submitted by authors:
Languages / tools used (tags)
Why you should attempt the paper.
No. attempts No. times reproduction has been attempted
Mean Repro Score Mean reproducibility score (out of 10)
- lower == harder!
Register paper using template in hackpad:
### **Paper:** <Title of the paper reproduced> **Reviewers:** Reviewer 1, Reviewer 2 etc.
Review as an auditor 📑
🔍 For FAIR materials
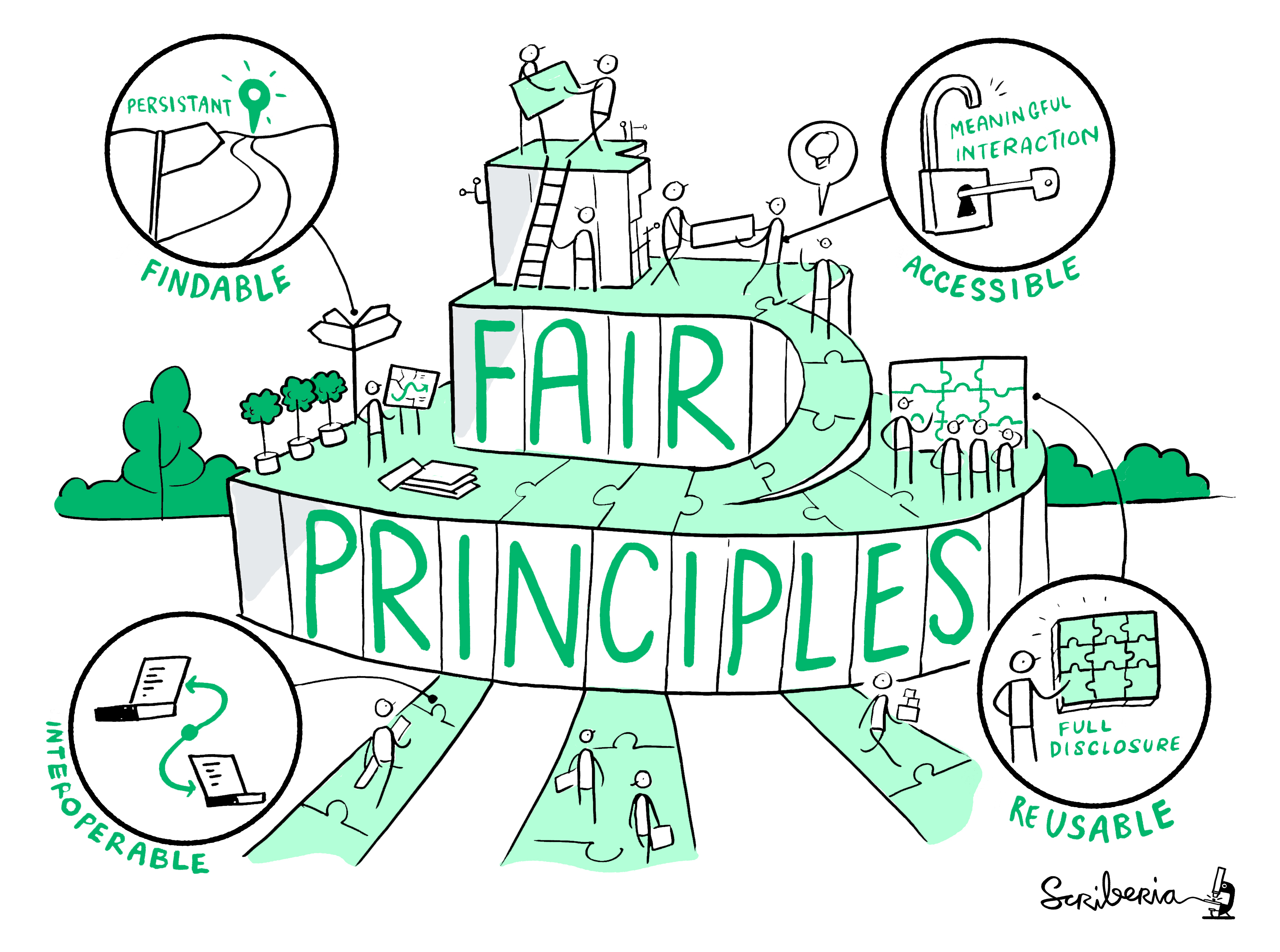
How easy was it to gain access to the materials?
Did you manage to download all the files you needed?
How easy / automated was installation?
Did you have any problems?
How did you solve them?
Were data clearly separated from code and other items?
Were large data files deposited in a trustworthy data repository and referred to using a persistent identifier?
Were data documented …somehow…
Was there adequate documentation describing:
how to install necessary software including non-standard dependencies?
how to use materials to reproduce the paper?
how to cite the materials, ideally in a form that can be copy and pasted?
Were you able to fully reproduce the paper?
✅
How automated was the process of reproducing the paper?
How easy was it to link analysis code to:
- the plots it generates
- sections in the manuscript in which it is described and results reported
🚫
Were there missing dependencies?
Was the computational environment not adequately described / captured?
Was there bugs in the code?
Did code run but results (e.g. model outputs, tables, figures) differ to those published? By how much?
Review as a user 🎮
Useful User Perspectives
New User

Invested User

What did you find easy / intuitive?
Was the file structure and file naming informative / intuitive? Was the analysis workflow easy to follow? Was there missing / confusing documentation?
What did you find confusing / difficult
Identify pressure points. Constructive suggestions?
What did you enjoy?
Identify aspects that worked well.
Feedback 💬
Feedback as a community member
Acknowledge author effort
Give feedback in good faith
Focus on community benefits and system level solutions
Help build convention on what a Research Compendium should be and how we should be able to use it

tl;dr: Don’t be this guy!

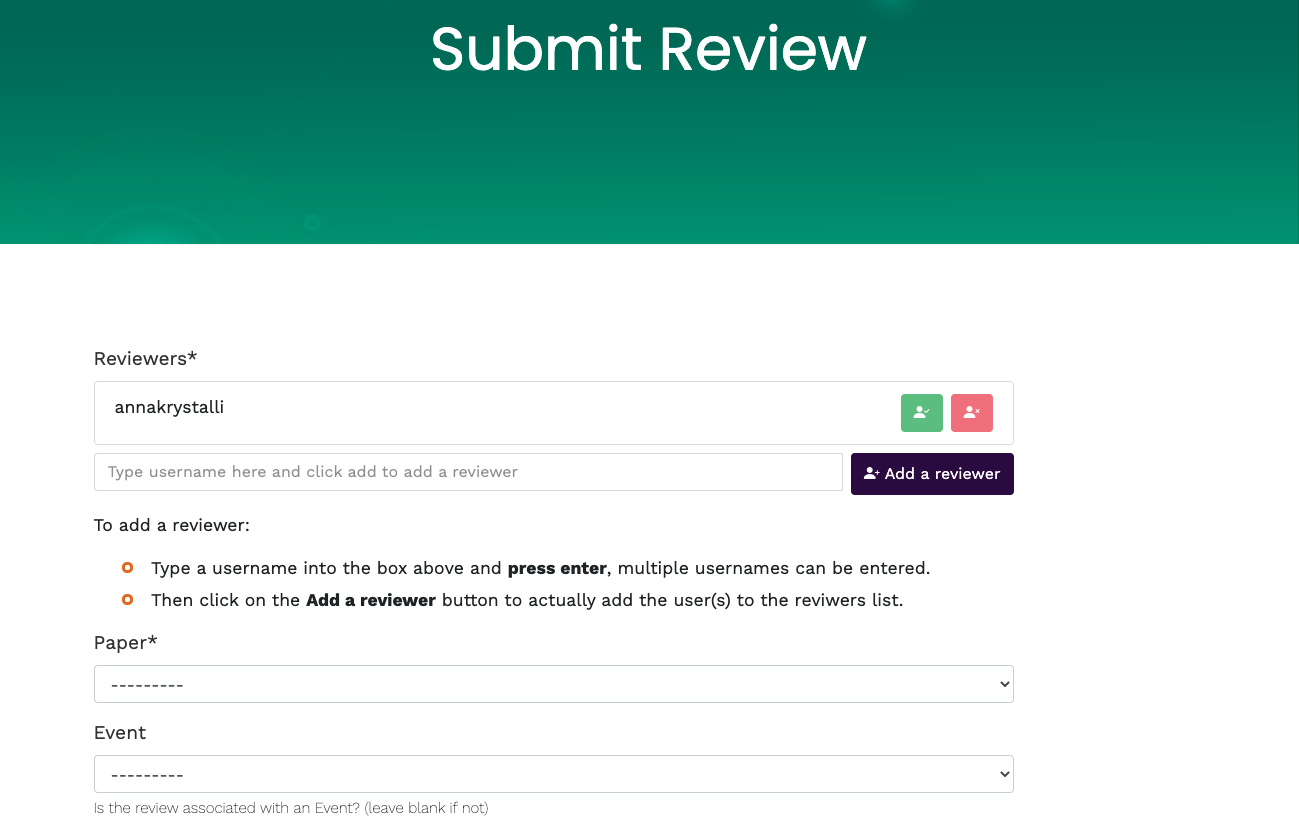
Submit review
Sign up / Log in
New Review: reprohack.org/review/new
Participant Guidelines
reprohack.org/participant_guidelines
Let’s go! 🏁
11:10 - 11:30
🔎 Paper List review
- Have a look at the papers available for reproduction
👥 Team formation / project registration
- Fine to work individually
- Add your details to the hackpad: bit.ly/mheim-reprohack-hackpad.
- Register your team and paper on the hackpad: bit.ly/mheim-reprohack-hackpad
🏠☕ Grab a coffee!
11:30 - 12:30 💻 ReproHack I
Work on your papers. Feel free to discuss papers and collaboratively troubleshoot problems.
Before Lunch-time Regroup 💭
Summarise group experiences
- What approaches to reproducibility the papers taken.
- Anything in particular you like about the approaches so far?
- Anything you’re having difficulty with?
12:30 - 12:45 💬 Lunch regroup
Feedback group experiences
12:45 - 13:45 🥗🌯 LUNCH
Lunch is just outside!
13:45 - 16:00 💻 ReproHack II & III
14:45 - 15:00 COFFEE BREAK 🏠☕
Work on your papers. Feel free to discuss papers and collaboratively troubleshoot problems.
Before Final-time Regroup 💭
Complete author feedback form ✍️
- Discuss how you got on with your papers?
- Summarise final experiences of the group in hackpad
TALK
📢 Camille Landesvatter
Research associate at the Mannheim Center for European Social Research (MZES)
‘Writing reproducible manuscripts in R Markdown and Pagedown’
16:25 - 16:50 Final regroup 💬
So, how did the groups get on?
Final comments.
On hackpad: Feedback
- One thing you liked
- One thing that can be improved.
Closing Remarks
Resources
The Turing Way: a lightly opinionated guide to reproducible data science.
Statistical Analyses and Reproducible Research: Gentleman and Temple Lang’s introduction of the concept of Research Compendia
Packaging data analytical work reproducibly using R (and friends): how researchers can improve the reproducibility of their work using research compendia based on R packages and related tools
How to Read a Research Compendium: Introduction to existing conventions for research compendia and suggestions on how to utilise their shared properties in a structured reading process.
Reproducible Research in R with rrtools: Workshop: Create a research compendium around materials associated with a published paper (text, data and code) using
rrtools.- Example Compendium: Demo Research compendium.
Did you enjoy ReproHacking? Get involved!
reprohack.org
Chat to us: ![Slack]()
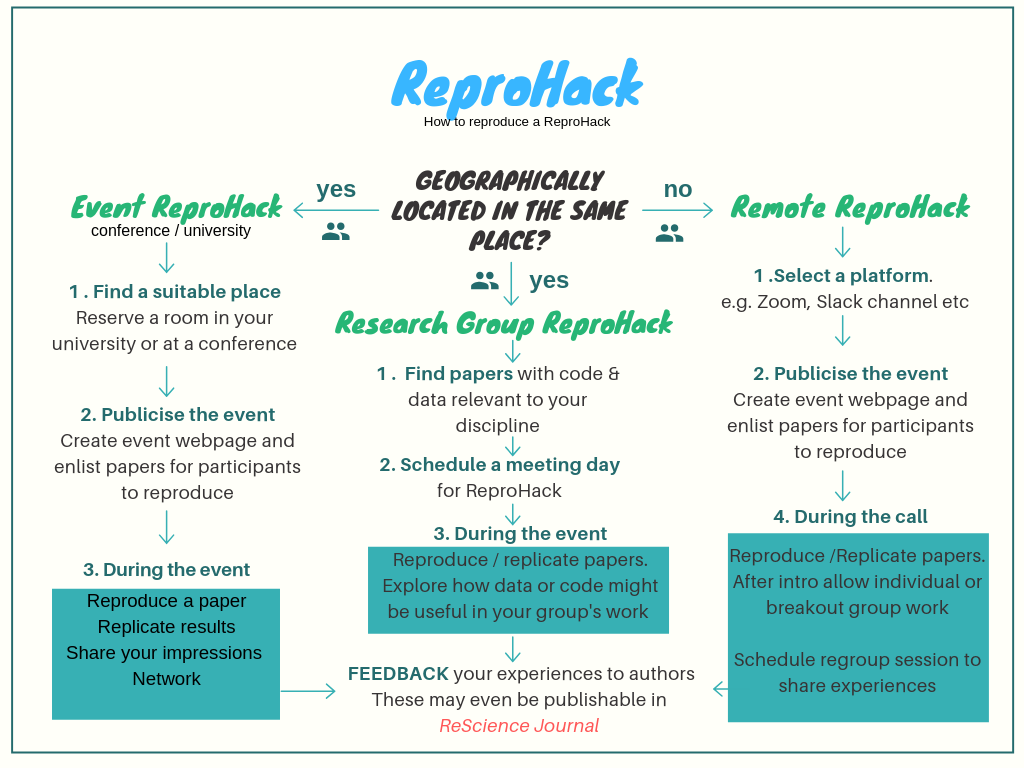
Host your own event!
Look out for train-the-trainer events!
Submit your own papers!
Many ways to ReproHack!
THANK YOU ALL! 🙏
Thank you PARTICIPANTS for coming!
Thank you AUTHORS for submitting!
Thank to the UoMannheim Open Science Office for sponsoring!
👋
Acknowledgements
Images throughout the slides watermarked with Scriberia were created by Scriberia for The Turing Way community and is used under a CC-BY licence:
- The Turing Way Community, & Scriberia. (2019, July 11). Illustrations from the Turing Way book dashes. Zenodo. http://doi.org/10.5281/zenodo.3332808

reprohack.org #MannheimReproHack #OSD22Mannheim